24
June
2020
E-CAS: Researchers Study The Brain’s Neural Code by Simulating Cortical Circuits on 100k Simultaneous Cores Using Google Cloud
Estimated reading time: 5 minutes
Authors: Salvador Durá-Bernal, Assistant Professor, State University of New York Downstate Health Sciences University, and Research Scientist IV, Nathan Kline Institute for Psychiatric Research; William W Lytton, Distinguished Professor, State University of New York.
Researchers from the State University of New York (SUNY) Downstate Health Sciences University presented the Phase 1 results of the NSF-funded Exploring Clouds for the Acceleration of Science (E-CAS) project to a scientific panel and the general public. This online presentation describes their efforts to decipher the brain’s neural code through large-scale detailed simulation of cortical circuits. To achieve this they developed novel methods and exploited state-of-the-art supercomputing technologies on Google Cloud Platform (GCP), including running simulations on 100k cores simultaneously.
Unlocking the mysteries of the brain has become one of the major challenges for humanity in the 21st century. Understanding the brain’s neural code is likely to have a profound and transformative impact, as it would help to develop treatments for brain disorders — depression, anxiety, Alzheimer’s, schizophrenia, Parkinson’s, epilepsy, amnesia, etc. — which the CDC estimates affect 1 out of 2 people. Potential treatments include targeted drugs and neurostimulation, as well as advancing brain-machine interfaces (BMI) to enable people with paralysis to control and feel using artificial limbs. Brain research would also help develop novel artificial intelligence (AI) algorithms, such as the revolutionary deep learning algorithm, which were derived from the function of neurons in the visual cortex.
Despite the large amount of experimental data, understanding the brain is challenging due to its complex interactions across a wide range of scales: from molecules to cells to circuits to behavior. Large-scale biophysically-detailed brain simulations provide an unrivaled method to integrate these data and bridge the scales (Fig. 1). However, this approach requires vast computational resources to run thousands of virtual experiments, each simulating the electrical currents of thousands of neurons and millions of synapses in a brain circuit.
Phase 1 of the E-CAS project enabled us to develop enhanced cloud computing methodologies and workflows. With the GCP-Slurm integration we were able to run large parameter-space explorations using up to 100k simultaneous cores, optimize large networks using evolutionary algorithms, and run long-duration simulations over 10 days. Placement groups enabled faster, low-latency simulations across multiple nodes. GCP Kubernetes enabled auto-scaling multi-user clusters to host our modeling application.
These cloud resources and improved workflows resulted in increased research productivity and scientific advances. We extended our mouse motor cortex model to include two new cell types, reproduced in vivo experimental data, and performed virtual experiments providing insights into oscillatory dynamics, molecule-to-behavior links critical for understanding drug effects, neuronal avalanches and information flow (Fig. 2). These resources also enabled building a novel detailed model of macaque auditory thalamocortical circuits, and provided access to a widely-accessible cloud-based tool for brain circuit modeling to the scientific community (http://netpyne.org).
The presentation describing our achievements enabled by E-CAS during this last year is available. The research was supported by the National Science Foundation (190444), the National Institutes of Health (U01EB017695, U24EB028998), and the New York State Department of Health (DOH01-C32250GG- 3450000).
Fig. 1: Web-based graphical interface of multiscale brain circuit modeling tool (NetPyNE). The tool is hosted on GCP and integrated with the Open Source Brain Platform (netpyne.opensourcebrain.org). It is free to access for all students researchers who can build, simulate and analyze brain circuit models online (www.netpyne.org). [1–4]
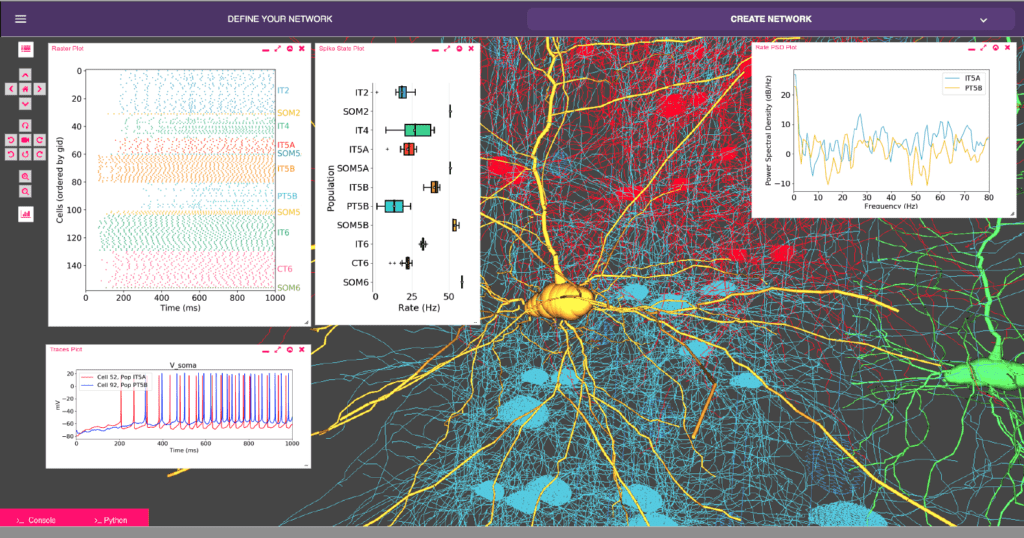
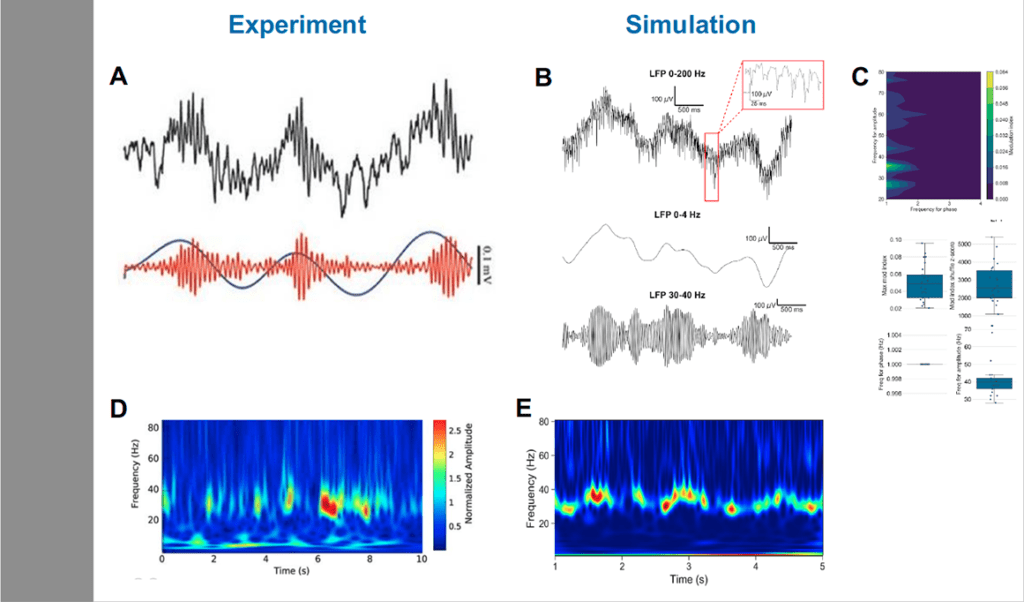
Fig. 2: Model reproduced oscillations and phase-amplitude coupling in cortical circuits. Experimental (A) and model (B) local field potential oscillations with delta and gamma frequencies showing phase-amplitude coupling. Phase-amplitude coupling modulation index demonstrating statistical validity across N=25 simulations (C). Experimental (D) and model (E) Spectrogram showing the power of delta and beta/gamma oscillations across time. [5–8]
References
- Dura-Bernal S, Suter BA, Gleeson P, Cantarelli M, Quintana A, Rodriguez F, et al. NetPyNE, a tool for data-driven multiscale modeling of brain circuits. Elife. 2019;8. doi:10.7554/eLife.44494
- Gleeson P, Cantarelli M, Marin B, Quintana A, Earnshaw M, Sadeh S, et al. Open Source Brain: A Collaborative Resource for Visualizing, Analyzing, Simulating, and Developing Standardized Models of Neurons and Circuits. Neuron. 2019. doi:10.1016/j.neuron.2019.05.019
- Dai K, Hernando J, Billeh YN, Gratiy SL, Planas J, Davison AP, et al. The SONATA data format for efficient description of large-scale network models. PLoS Comput Biol. 2020;16: e1007696.
- Alber M, Buganza Tepole A, Cannon WR, De S, Dura-Bernal S, Garikipati K, et al. Integrating machine learning and multiscale modeling-perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. NPJ Digit Med. 2019;2: 115.
- Dura-Bernal S, Neymotin SA, Suter BA, Shepherd GMG, Lytton WW. Multiscale dynamics and information flow in a data-driven model of the primary motor cortex microcircuit. BiorXiv 101101/201707. 2019. doi:10.1101/201707
- Neymotin SA, Suter BA, Dura-Bernal S, Shepherd GMG, Migliore M, Lytton WW. Optimizing computer models of corticospinal neurons to replicate in vitro dynamics. J Neurophysiol. 2016;117: jn.00570.2016.
- Neymotin SA, Dura-Bernal S, Lakatos P, Sanger TD, Lytton W. Multitarget multiscale simulation for pharmacological treatment of dystonia in motor cortex. Front Pharmacol. 2016;7: 157.
- Andino-Pavlovsky V, Souza AC, Scheffer-Teixeira R, Tort ABL, Etchenique R, Ribeiro S. Dopamine Modulates Delta-Gamma Phase-Amplitude Coupling in the Prefrontal Cortex of Behaving Rats. Front Neural Circuits. 2017;11: 29.
Related articles and blog posts:
